- Exon count:
- 20
| Annotation release |
Status |
Assembly |
Chr
|
Location |
|
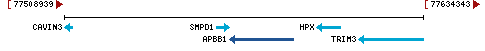
103
|
current |
EquCab3.0 (GCF_002863925.1) |
7 |
NC_009150.3 (77566689..77588780, complement) |
| 102 |
previous assembly |
EquCab2.0 (GCF_000002305.2) |
7 |
NC_009150.2 (75389177..75411415, complement) |
Chromosome 7 - NC_009150.3