?
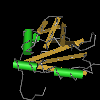 
Putative hydrophobic ligand-binding SRPBCC domain of the Hsp90 co-chaperone Aha1 and related proteins This subfamily includes the C-terminal SRPBCC (START/RHO_alpha_C/PITP/Bet_v1/CoxG/CalC) domain of Aha1, and related domains. Proteins in this group belong to the SRPBCC domain superfamily of proteins which bind hydrophobic ligands. SRPBCC domains have a deep hydrophobic ligand-binding pocket. Aha1 is one of several co-chaperones, which regulate the dimeric chaperone Hsp90. Hsp90, Aha1, and other accessory proteins interact in a chaperone cycle driven by ATP binding and hydrolysis. Aha1 promotes dimerization of the N-terminal domains of Hsp90, and stimulates its low intrinsic ATPase activity. One Aha1 molecule binds per Hsp90 dimer. The N- and C- terminal domains of Aha1 cooperatively bind across the dimer interface of Hsp90. The C-terminal domain of Aha1 binds the N-terminal Hsp90 ATPase domain. Aha1 may regulate the dwell time of Hsp90 with client proteins. Aha1 may act as either a negative or positive regulator of chaperone-dependent activation, depending on the client protein; for example, it acts as a negative regulator in the case of Saccharomyces cerevisiae MAL63 MAL-activator, and acts as a positive regulator in the case of glucocorticoid receptor and v-Src kinase. The mechanisms by which these opposing functions are achieved are unclear. Aha1 is upregulated in a number of tumor lines co-incident with the activation of several signaling kinases. |
